Hyper-parameter scans#
Here we consider how different optimization algorithms behave (for this system) and how their hyper parameters influence the resulting cluster expansion. We use the cutoffs [8.0, 6.5, 6.0] and consider the following optimization algorithms
ARDR (Automatic Relevance Determination Regression)
RFE (Recursive Feature Elimination)
LASSO (Least Absolute Shrinkage and Selection Operator)
Adaptive-LASSO
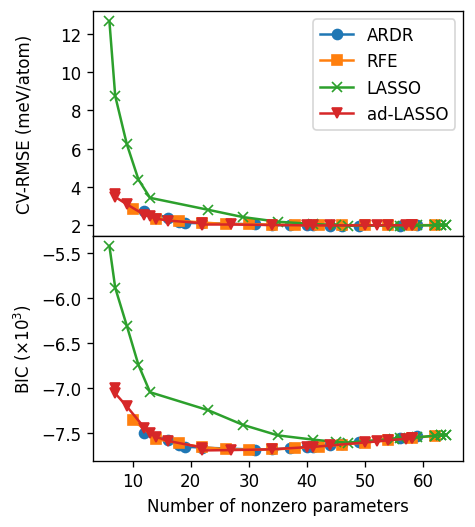
From the analyis above one can conclude that around 20 to 30 non-zero ECIs are a good choice with cutoffs [8.0, 6.5, 6.0]. We also note here that LASSO performs quite poorly compared to the other methods, whereas the other three methods all yield similar results.
[1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from ase.db import connect
from icet import ClusterSpace, StructureContainer
from trainstation import CrossValidationEstimator
Preparations#
We set up the cluster space and prepare the reference data.
[2]:
# access database with reference data
db = connect('../../tutorial/reference_data.db')
# set up cluster space
primitive_structure = db.get(id=1).toatoms()
cs = ClusterSpace(
structure=primitive_structure,
cutoffs=[8.0, 6.5, 6.0],
chemical_symbols=['Ag', 'Pd']
)
# compile fit data
sc = StructureContainer(cluster_space=cs)
for row in db.select():
sc.add_structure(
structure=row.toatoms(),
user_tag=row.tag,
properties={'mixing_energy': row.mixing_energy},
)
A, y = sc.get_fit_data(key='mixing_energy')
Furthermore we define a convenience function that extracts pertinent information from a CrossValidationEstimator object.
[3]:
def get_row(cve: CrossValidationEstimator):
row = dict()
row['rmse_validation'] = cve.rmse_validation
row['rmse_train'] = cve.rmse_train
row['BIC'] = cve.model.BIC
row['n_parameters'] = cve.n_parameters
row['n_nonzero_parameters'] = cve.n_nonzero_parameters
return row
ARDR: Hyperparameter scan#
ARDR has the hyper-parameter threshold-lambda which controls the sparsity of the solution.
[4]:
lambda_values = [250, 500, 1000, 1400, 2000, 2500, 4500, 7500,
13000, 18000, 25000, 40000, 60000, 90000, 200000]
records = []
for lam in lambda_values:
cve = CrossValidationEstimator((A, y), fit_method='ardr', threshold_lambda=lam)
cve.validate()
cve.train()
row = get_row(cve)
row['threshold_lambda'] = lam
records.append(row)
df_ardr = pd.DataFrame(records)
RFE: Hyperparameter scan#
RFE has the hyper-parameter n_features which controls the sparsity of the solution.
[5]:
nf_values = np.arange(10, len(cs), 4)
records = []
for nf in nf_values:
cve = CrossValidationEstimator((A, y), fit_method='rfe', n_features=nf)
cve.validate()
cve.train()
row = get_row(cve)
records.append(row)
df_rfe = pd.DataFrame(records)
LASSO: Hyperparameter scan#
[6]:
alpha_values = np.logspace(-5, -0.5, 20)
records = []
for alpha in alpha_values:
cve = CrossValidationEstimator((A, y), max_iter=50000, fit_method='lasso', alpha=alpha)
cve.validate()
cve.train()
row = get_row(cve)
row['alpha'] = alpha
records.append(row)
df_lasso = pd.DataFrame(records)
Adaptive-LASSO: Hyperparameter scan#
[7]:
alpha_values = np.logspace(-5, -1.5, 20)
records = []
for alpha in alpha_values:
cve = CrossValidationEstimator((A, y), max_iter=50000,
fit_method='adaptive-lasso', alpha=alpha)
cve.validate()
cve.train()
row = get_row(cve)
row['alpha'] = alpha
records.append(row)
df_adlasso = pd.DataFrame(records)
Plot results#
[8]:
fig, axes = plt.subplots(
figsize=(4, 4.5),
dpi=120,
sharex=True,
nrows=2,
)
ax = axes[0]
conv = 1e3
ax.set_ylabel('CV-RMSE (meV/atom)')
ax.plot(df_ardr.n_nonzero_parameters, conv * df_ardr.rmse_validation,
'-o', label='ARDR')
ax.plot(df_rfe.n_nonzero_parameters, conv * df_rfe.rmse_validation,
'-s', label='RFE')
ax.plot(df_lasso.n_nonzero_parameters, conv * df_lasso.rmse_validation,
'-x', label='LASSO')
ax.plot(df_adlasso.n_nonzero_parameters, conv * df_adlasso.rmse_validation,
'-v', label='ad-LASSO')
ax.legend()
ax = axes[1]
conv = 1e-3
ax.set_ylabel(r'BIC ($\times 10^{3}$)')
ax.plot(df_ardr.n_nonzero_parameters, conv * df_ardr.BIC, '-o', label='ARDR')
ax.plot(df_rfe.n_nonzero_parameters, conv * df_rfe.BIC, '-s', label='RFE')
ax.plot(df_lasso.n_nonzero_parameters, conv * df_lasso.BIC, '-x', label='LASSO')
ax.plot(df_adlasso.n_nonzero_parameters, conv * df_adlasso.BIC, '-v', label='ad-LASSO')
axes[-1].set_xlabel('Number of nonzero parameters')
fig.tight_layout()
fig.subplots_adjust(hspace=0)
fig.align_ylabels(axes)